Transient transfection refers to the introduction of plasmids or other vectors carrying exogenous DNA into cells, where they exist in a non-integrated form and express the contained genes for a limited period. This approach is often used for short-term gene expression studies and can be achieved using standard plasmid vectors like pcDNA3.1 and pUC57.
In contrast, stable transfection involves integrating the exogenous gene into the host cell's genome to achieve long-term and stable gene expression. Constructing stable cell lines requires gene editing technologies, which primarily include the following two techniques:
DNA Transposon Technology: In nature, DNA transposon systems include transposase, an enzyme responsible for recognizing and catalyzing the movement of transposons within the genome. Transposons have terminal inverted repeat sequences (TRs) at both ends. Transposase can introduce double-strand breaks at the TRs and genomic sites, facilitating the transposition of the entire transposon region. For gene editing applications, transposase components can be separated into:
1. A helper plasmid encoding the transposase.
2. A transposon plasmid containing optimized TRs at both ends, with the transposon region in between where the gene of interest can be inserted.
Commonly used transposon systems include PiggyBac, Sleeping Beauty, and Tol2, each with its specific characteristics as detailed in the table below.
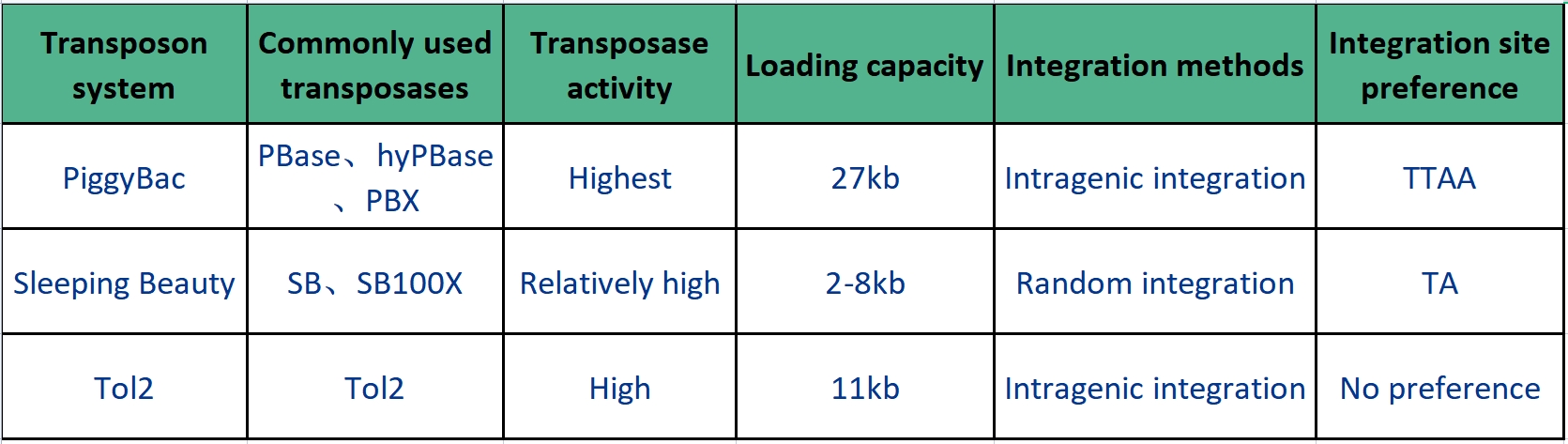
Table 1: Characteristics of PiggyBac, Sleeping Beauty, and Tol2
Among them, PiggyBac is one of the most popular transposon systems, used for delivering transgenes, shRNA, recombinant antibodies, and more. Since its discovery, PiggyBac transposons have undergone a series of optimizations and modifications. The optimized transposase (hyPBase) enhances transposition efficiency, with cutting activity but lacking integration activity. This ensures that once the integrated sequence is removed, the position will not be reinserted by the transposon, achieving seamless cutting.
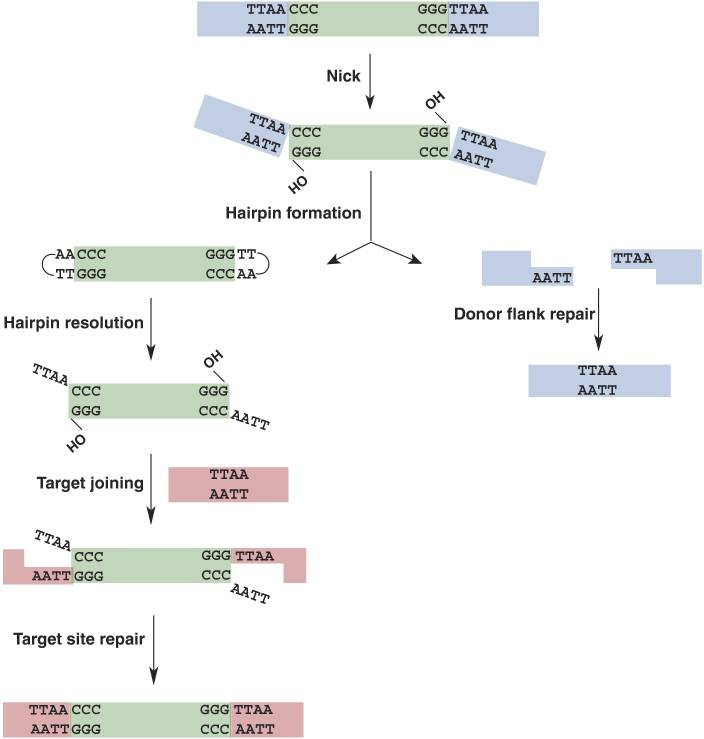
Additionally, a significant advantage of the PiggyBac transposon is its large cargo capacity, able to carry up to 27 kb of DNA between the 5' and 3' terminal repeats (TRs). During gene integration mediated by PiggyBac transposons, DNA is cut at the chromosomal TTAA sites, with a preference for integrating into gene regions. The specific integration process for the PiggyBac system is illustrated in the figure below:
Figure 1: PiggyBac Transposon Cleavage and Repair Mechanism
CRISPR/Cas9 Technology: CRISPR/Cas9 is a type II CRISPR system. It only requires one protein, the Cas9 protein, for the cleavage of foreign genes. Due to its straightforward design and operation, it has been widely used in gene editing to achieve gene knockout, knock-in, and transcriptional activation. When constructing a gene knockout stable cell line, the process begins with plasmid design, which includes a guide RNA (sgRNA, targeting the gene of interest), the Cas9 enzyme gene, and an EGFP or other marker protein. After plasmid transfection into cells, gene editing occurs within the cell: guided by the sgRNA, the Cas9 nuclease cleaves the DNA double strand at a specific position adjacent to the PAM sequence (i.e., at the 5' end), resulting in double-strand breaks in the DNA.
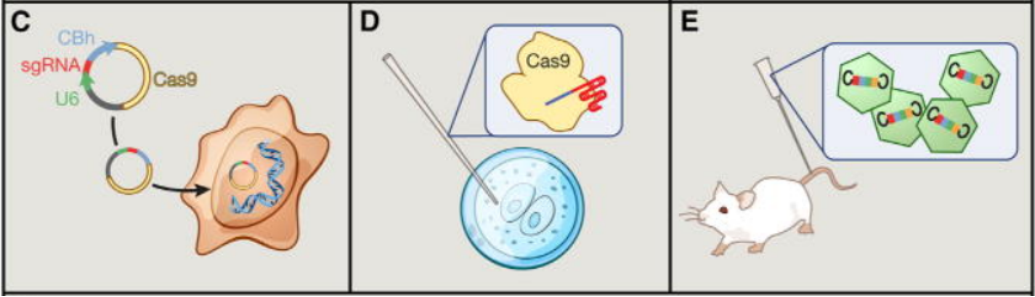 Figure 2:Application of Cas9 as a Gene Editing Tool in Cells (Left: Cell Lines; Center: Zygotes; Right: Somatic Cells)
Figure 2:Application of Cas9 as a Gene Editing Tool in Cells (Left: Cell Lines; Center: Zygotes; Right: Somatic Cells)
2. Lentivirus-Mediated Cell Infection:
Direct plasmid transfection and selection is one of the earliest methods used for constructing cell lines, but it has relatively low efficiency and poor stability. In contrast, lentiviruses can infect almost all types of cells (both dividing and non-dividing) and efficiently integrate genetic material into the host cell genome, achieving long-term stable expression. Due to its high infection efficiency and ease of use, lentivirus-mediated methods have become the mainstream technology for constructing stable cell lines. The construction of stable cell lines using lentivirus involves three main steps: constructing the lentivirus expression vector, packaging the lentivirus, and establishing the stable cell line.
Ⅲ. Advantages of Stable Cell Lines
1.Long-Term Stable Expression: Stable cell lines can express foreign genes consistently over long periods, facilitating extended experiments and research, with most research cell lines capable of stable, repeated passaging.
2. Reproducibility and Accuracy: The consistent genetic background of stable cell lines provides high experimental reproducibility and accuracy of results.
3. Functional Research: Stable cell lines are used for in-depth studies of specific gene functions by continuously expressing the target gene, allowing researchers to investigate its role in cellular physiological and pathological processes.
4. Gene Editing Validation: Stable cell lines are useful for validating gene editing techniques, such as CRISPR/Cas9, by evaluating editing efficiency through stable expression of reporter genes. They can also be used to achieve gene knockout, knock-in, transcriptional activation, or repression for functional exploration of genes.
5. Disease Model Construction: Stable cell lines can simulate gene expression changes related to diseases, helping in the construction of disease models and providing a platform for studying disease mechanisms and developing therapeutic strategies.
6. Drug Screening and Development: In drug screening, stable cell lines serve as model systems for high-throughput screening and evaluation of compound efficacy and safety.
7. Production of Biopharmaceuticals: In the biotechnology industry, stable cell lines are used for the production of recombinant proteins, antibodies, and other biopharmaceutical products.
Brain Case Biotech offers a complete suite of stable cell line construction services (including transposon and lentivirus methods). If you have any needs related to stable cell line development, please feel free to contact us for more details or to place a custom order!
References:
[1]Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008 Apr 9;27(7):1097-109.
[2]Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014 Jun 5;157(6):1262-1278.
[3]Wu Y, Wang J, Zhao T, Chen J, Kang L, Wei Y, Han L, Shen L, Long C, Wu S, Wei G. Di-(2-ethylhexyl) phthalate exposure leads to ferroptosis via the HIF-1α/HO-1 signaling pathway in mouse testes. J Hazard Mater. 2022 Mar 15;426:127807.
[4]Yu L, Sui B, Zhang X, Liu J, Hao X, Zheng L. miR-92a-1-5p enriched prostate cancer extracellular vesicles regulate osteoclast function via MAPK1 and FoxO1. J Exp Clin Cancer Res. 2023 May 2;42(1):109.
If you are interested in the details of the experiment or possible problems and causes during the experiment, please contact: BD@ebraincase.com