- E-mail:BD@ebraincase.com
- Tel:+8618971215294
GRASP (GFP Reconstitution Across Synaptic Partners) uses green fluorescent protein GFP to detect the distance between neurons and whether synaptic connections are formed. It was developed by Kang Shen, a Chinese scientist at Stanford University and Cornelia I. Bargmann of Rockefeller University in the United States. This technology can achieve synaptic positioning in the living nervous system. To put it simply, GFP is divided into two components, and the separated GFP components do not have the function of emitting light. The two components of GFP are expressed in two groups of neurons respectively. When there is a synaptic connection between the two groups of neurons, the two components of GFP automatically combine at the synaptic site to form a complete functional GFP. At this time, the synaptic structure is generated. Green fluorescence. If there are no synaptic connections between the two groups of neurons, no fluorescence is produced.
In 2011, researchers from the Howard Hughes Medical Institute, Korea Advanced Institute of Science and Technology, Zhejiang University and other institutions published a research paper titled mGRASP enables mapping mammalian synaptic connectivity with light microscopy in Nature Methods. The vector was transformed and the mammalian GRASP (mGRASP) technology was developed, which can realize the positioning of neuron synapses in the mouse brain and realize the rapid and accurate reconstruction of the mammalian brain neural network under an optical microscope. Through mGRASP, we can observe the changes in neural networks under these diseases and infer their pathological mechanisms.
Cre-independent pre-mGRASP (rAAV CAG-pre-mGRASP-mCerulean) is expressed in CA3 neurons of the left hemisphere of mice, and Cre recombinase-dependent post-mGRASP (rAAV CAG-Jx-rev- post-mGRASP-2A-dTomato). Neither the split-GFP fragment fluoresces when expressed alone, but mGRASP is reorganized across synapses, at locations where mCerulean-labeled CA3 axons and dTomato-labeled CA1 dendrites intersect, along the dTomato-labeled CA1 apical and basal trees Discrete fluorescent spots were highlighted (Fig. 1B,C).
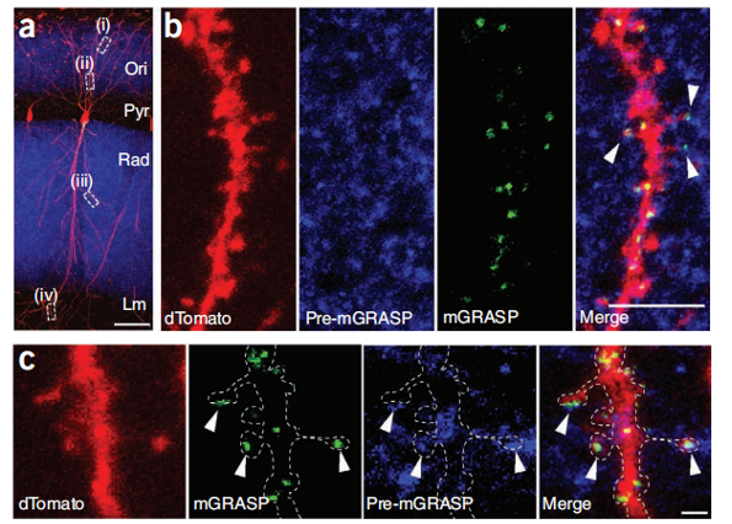
Figure 1: mGRASP reconstruction of hippocampal CA3-CA1 connectivity. (A) Discrete spots of recombinant mGRASP fluorescence in single CA1 neurons. (B,C) Cropped high-magnification images showing reconstructed mGRASP signal where blue axons intersect with red dendrites (arrows). Scale bars, 5 µm (B) and 1 µm (C).
In 2014, researchers from the Howard Hughes Medical Institute, Korea Advanced Institute of Science and Technology, Zhejiang University and other institutions published a paper in Nature Protocols entitled Using mammalian GFP reconstitution across synaptic partners (mGRASP) to map synaptic connectivity in the mouse brain research paper, the authors introduce several mGRASP variants with Cre dependence and fluorescence color selection to allow selection in experimental strategies. These mGRASP viral constructs, combined with a large set of Cre lines, enable the selection and induction of synaptic markers, allowing the study of synaptic connectivity patterns in many types of neural circuits, such as cell type-specific local and long-range connections (Fig. 2).
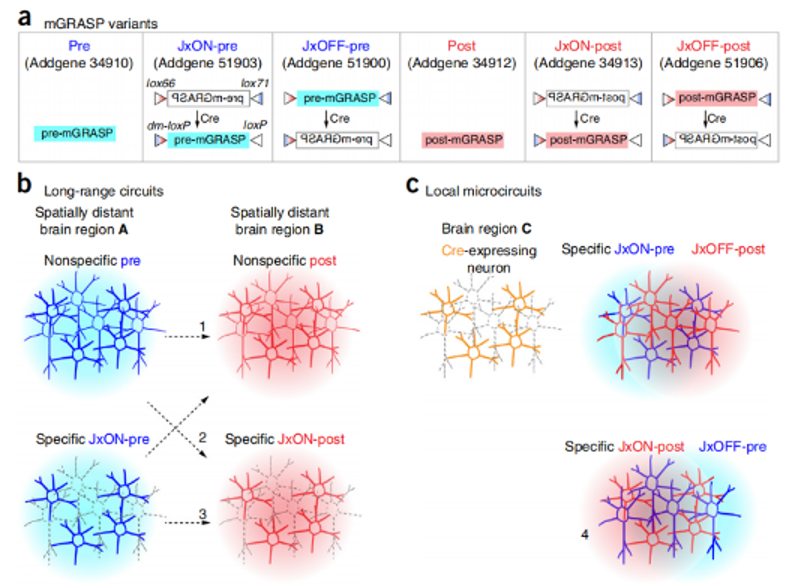
Figure 2: Schematic of mGRASP variants and experimental design of mGRASP auxiliary circuit mapping. (A) Schematic representation of mGRASP variants. (B,C) Various long-range (B) and local circuits (C) can be mapped by injecting mGRASP AAV variants into brain regions of interest in wild-type (non-specific) or cell type-specific Cre mouse lines.
In 2018, researchers from the School of Biological Sciences at Seoul National University published a research paper titled Interregional synaptic maps among engram cells underlie memory formation in Science, in order to compare the projection of two different presynaptic populations in neurons to a single postsynaptic Neuron, the authors improved the GRASP technology and developed an enhanced GRASP (eGRASP) technology that promotes GFP reconstruction by introducing a weak interaction domain, enhances the GRASP signal intensity, and further evolves eGRASP to reconstruct cyan or yellow fluorescent protein (Figure 3A,B). Placing the color-determining domain on the presynaptic neuron (cyan/yellow pre-eGRASP) and the common domain on the postsynaptic neuron (post-eGRASP) allows visualization of two synaptic populations that From two different presynaptic neuron populations and projecting to a single postsynaptic neuron, the authors named this technique dual-eGRASP (Figure 3A). The authors successfully applied this technique to synapses on dentate gyrus (DG) granule cells derived from the lateral entorhinal cortex (LEC) or medial entorhinal cortex (MEC), respectively, which project to the outer layers of the DG and the middle molecular layer (Fig. 3D).

Figure 3: Dual-eGRASP differentiates between two synaptic populations on a single neuron. (A,B) Schematic diagram of cyan and yellow eGRASP. Cyan pre-eGRASP and yellow pre-eGRASP are expressed in two different presynaptic cells, while ordinary post-eGRASP is expressed in a single postsynaptic cell. (D)) LEC and MEC express cyan and yellow pre-eGRASP, respectively, and post-eGRASP is expressed together with myristoylated TagRFP-T (myr_TagRFP-T) in DG.
mGRASP was used to test whether enhanced MD synaptic input in PTSD mice remodels local synaptic connections in the ACC. This technology is based on the expression of two functionally complementary non-fluorescent GFP fragments (AAV2/8-CAG-pre-GRASP-mCerulean AAV2/8-CAG-DIO-post-GRASP-P2A- tdTomato, provided by Braincase), specifically labels synapses between the two neuronal populations, MD and ACC, for more accurate and specific visualization (Figure 4M). Experimental results show that the thalamocortical synaptic frequency of PV+ neurons in the ACC area of WT mice is significantly higher than that of pyramidal neurons (that is, CaMKII+ neurons), which means that there may be a stronger connection between MD and ACC PV+ neurons (Figure 4N,O). The number of thalamocortical synapses on PV+ interneurons was increased in PTSD mice compared with WT mice (Fig. 4Q,S). There was no significant difference in the synapses of pyramidal neurons between the two groups of mice (Figure 4P,R). This shows that during the fear extinction process in PTSD mice, MD preferentially projects to the PV+ neurons of the ACC.
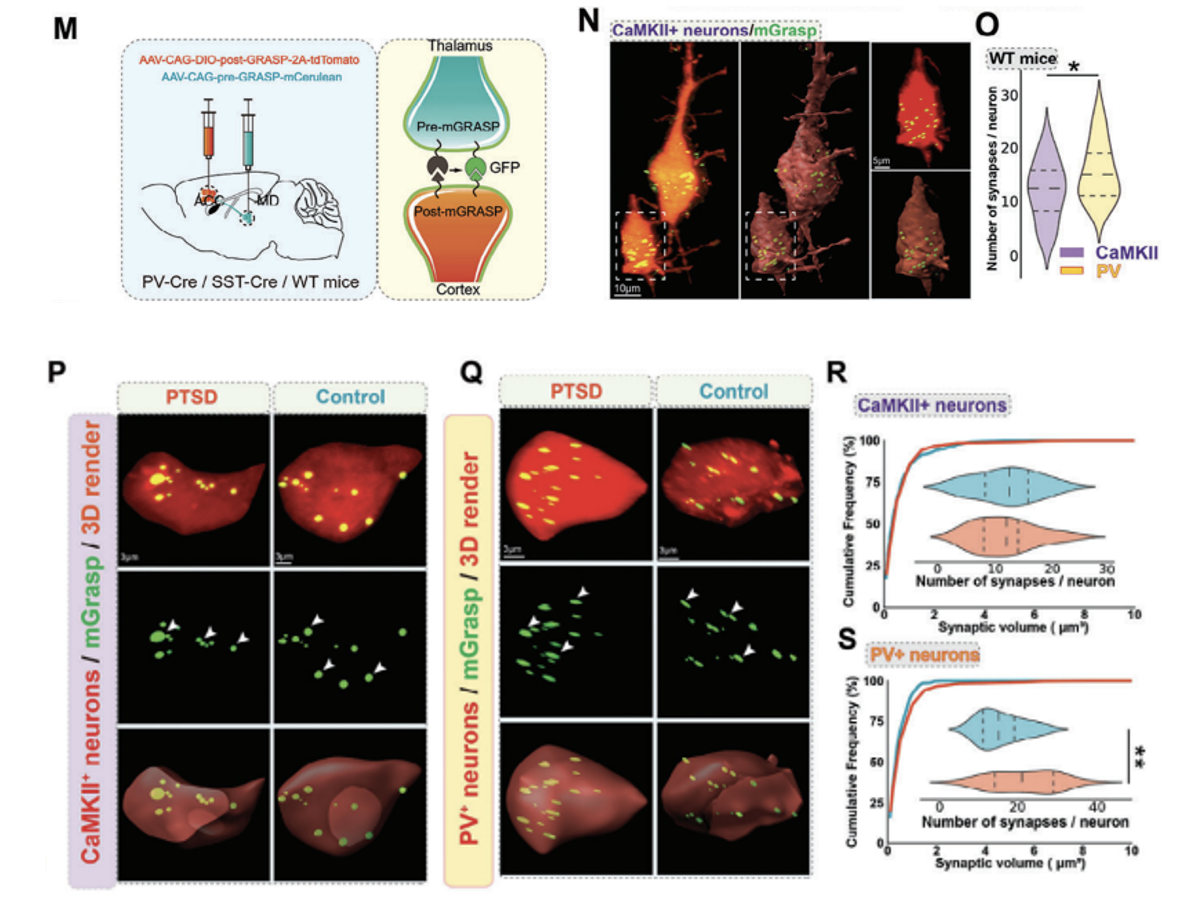
Figure 4: MD preferentially projects to PV+ neurons in the ACC during fear extinction in PTSD mice. (M) Schematic of the mechanism for viral labeling of synapses using PV-Cre, SST-Cre, or CaMKII-Cre. (N, O) High-magnification confocal image (N) and quantification map (O) of the synaptic frequency of CaMKII+ and PV+ neurons in the thalamocortex of WT mice. (P,Q) High-power confocal (top) and 3D images (middle, bottom) of mGRASP-labeled CaMKII+ neurons (P)/PV+ neurons (Q) and synapses (green) in the ACC. (R,S) Quantification of synaptic frequency of CaMKII+ neurons (R)/PV+ neurons (S) in the thalamocortex of WT mice and PTSD mice.
By further modifying the vector, the researchers developed mammalian GRASP (mGRASP) and enhanced dual-GRASP technology (dual-eGRASP), which can not only achieve the positioning of neuronal synapses in the mouse brain, but also combine chemical technology with computer bioimage informatics technology enables high-throughput neural network reconstruction and can be used for the study of neurological diseases. For example, neurological diseases such as depression, Parkinson, and epilepsy may have abnormal connections of neurons. These can be observed through GRASP Changes in neural networks under disease to infer pathological mechanisms.
| # | Product type | Product number | Product name |
|---|---|---|---|
| 1 | Fluorescent labeling | BC-1424 | rAAV-EWB-DIO-myriRFP670V5-P2A-post-Egrasp |
| 2 | Fluorescent labeling | BC-1425 | rAAV-CWB-pre-eGRASP(p30) |
| 3 | Fluorescent labeling | BC-1626 | rAAV-CAG-pre-mGRASP-mCerulean |
| 4 | Fluorescent labeling | BC-1627 | rAAV-CAG-DIO-post-mGRASP-T2A-dTomato |
| 5 | Fluorescent labeling | BC-2371 | rAAV-CAG-DIO-pre-mGRASP-miRFP670nano3 |
| 6 | Fluorescent labeling | BC-2372 | rAAV-CAG-post-mGRASP-T2A-dTomato |