- E-mail:BD@ebraincase.com
- Tel:+8618971215294
In 1981, Nat Sternberg et al. reported a protein cloned from the P1 bacteriophage of Escherichia coli—a 38 kDa recombinase encoded by 343 amino acids called Cre (Cyclization Recombination Enzyme). It specifically recognizes loxP sites and mediates sequence deletion or recombination between loxP sites. The loxP (locus of X-overP1) site is 34 bp long, consisting of two 13 bp inverted repeat sequences and an 8 bp spacer region. The inverted repeat sequences are the specific recognition sites for the Cre recombinase, while the spacer region determines the orientation of loxP.
The result of DNA recombination depends on the direction and position of the two loxP sites, primarily leading to the following types of recombination:
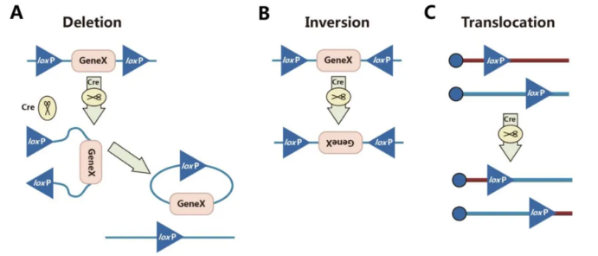
A. Deletion: When two loxP sites are on the same DNA strand and oriented in the same direction, the Cre recombinase excises the sequence between the loxP sites.
B. Inversion: When two loxP sites are on the same DNA strand and oriented in opposite directions, the Cre recombinase inverts the sequence between the loxP sites.
C. Translocation: When two loxP sites are on different DNA strands or chromosomes, the Cre recombinase induces an exchange between the DNA strands or a chromosomal translocation.
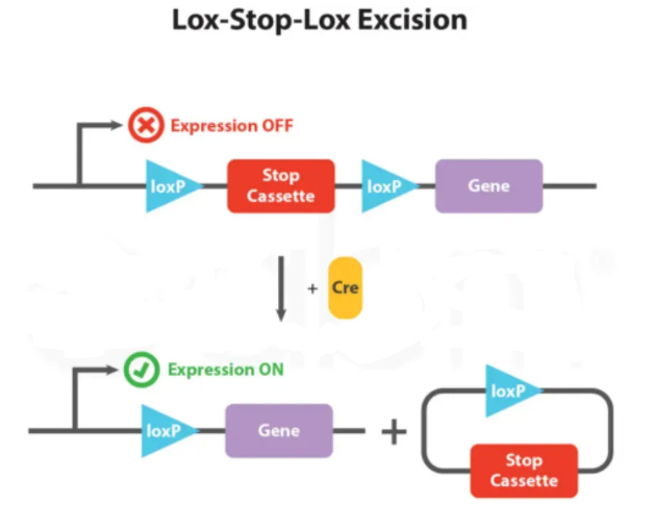
Based on the above principles, the LSL sequence (loxP-STOP-loxP) was designed. In the absence of Cre recombinase, the downstream gene cannot be normally expressed due to the presence of the STOP cassette. Upon introduction of Cre recombinase, the STOP sequence is deleted, allowing the downstream gene to be expressed normally, thereby achieving Cre-dependent gene expression.
However, in practical applications, it was found that LSL could still lead to slight expression of the downstream gene even in the absence of Cre recombinase. In 2003, Frank Schnütgen et al. developed FLEX (Flip-excision) and, based on this, DIO (Double-Floxed Inverted Open reading frame), which changes the orientation of loxP and lox2272, greatly mitigating the leakage issue. lox2272 is also a commonly used lox site, similar to loxN, lox511, etc. lox2272 and loxP are mutually incompatible and are respectively targeted by Cre recombinase. In the DIO structure, Cre recombinase causes one inversion and one deletion through loxP and lox2272, thus achieving irreversible inversion and Cre-dependent expression of the target gene.
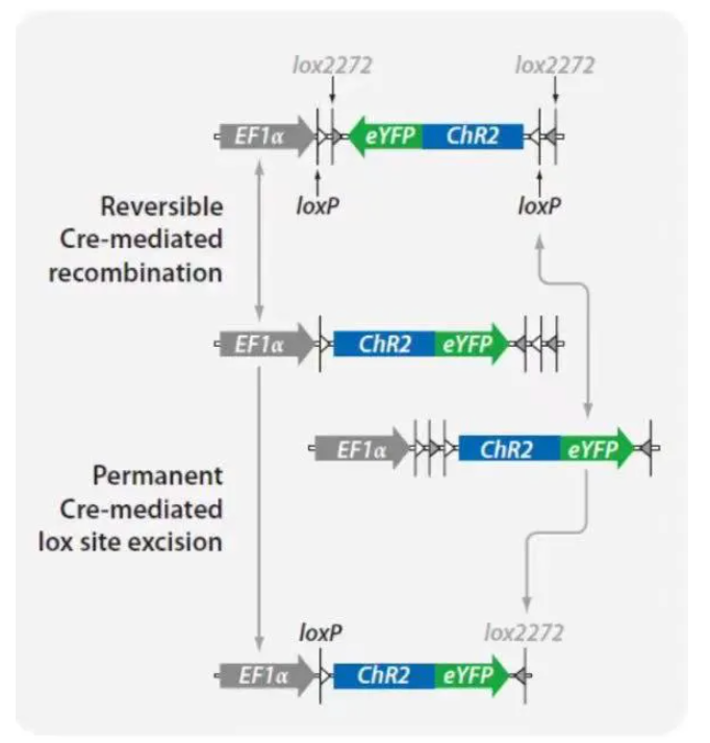
The inserted gene is oriented in the same direction as the promoter. In the presence of Cre recombinase, recombination occurs, causing the gene to invert and thus preventing normal expression.
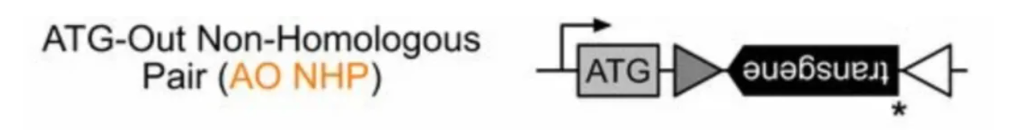
Given the leakage expression issues with both LSL and DIO/FLEX, the Edward M. Callaway team designed a CIAO (cross-over insensitive ATG-Out) sequence, which almost eliminates the leakage problem. CIAO uses the ATG-OUT construction method, placing the ATG and the ORF of the target gene on opposite sides of the lox sequence. This means that if recombination does not occur, the target gene is incomplete, and even if transcription of the gene occurs, no functional protein will be translated. Additionally, the non-symmetrical lox66/71 sequences are used to further reduce spontaneous recombination during plasmid replication in bacteria. However, compared to loxP and lox2272, the recombination efficiency of Cre recombinase with lox66/71 is also reduced.
This is because using viruses to achieve target gene overexpression (OE) or knockdown (KD) often results in a shorter experimental period and lower experimental costs compared to mouse breeding. Additionally, OE/KD resulting from mouse breeding is generally systemic, whereas viruses can more easily achieve gene regulation in local tissues or specific organs, allowing for more detailed studies.
| # | Cre mice x Floxed mice | Cre mice x DIO Virus | Floxed mice x Cre Virus |
|---|---|---|---|
| CKO | One of the most commonly used and stable methods for gene knockout | Injecting AAV (carrying ShRNA or SgRNA/Cas9) into Cre mice can achieve local or systemic knockout | Injecting AAV-Cre into floxed mice can achieve local or systemic knockout |
| CKI | Commonly used reporter mice include mT/mG, Ai9, Ai14, etc | Injecting AAV-DIO-GOI into Cre mice can achieve local or systemic overexpression | Specific cell labeling can be achieved by injecting Cre virus into reporter mice |
iCre improves expression and activity in mammalian cells by optimizing Cre codons for mammalian preferences.
CreER is a fusion protein of Cre recombinase and the estrogen receptor (ER). Without an inducer, the expressed CreER binds to HSP90 protein and is localized in the cytoplasm, preventing it from entering the nucleus to edit genomic DNA. Only when an inducer (Tamoxifen or 4-OHT) is added, the inducer binds to the CreER protein, causing a conformational change that releases CreER from the anchoring protein HSP90. This allows CreER to enter the nucleus, recognize loxP sites, and induce recombination. By controlling the timing of inducer injection, temporal specificity of gene recombination can be achieved.
To avoid interference from endogenous estrogen, a point mutation (G521R) was introduced in the ligand-binding domain of the estrogen receptor, resulting in CreERT, which responds only to synthetic exogenous estrogens (Tamoxifen or 4-OHT). CreERT2 is a further improved version of CreERT, containing three additional point mutations (C400V/M453A/L544A), making it more sensitive and faster in response to Tamoxifen.
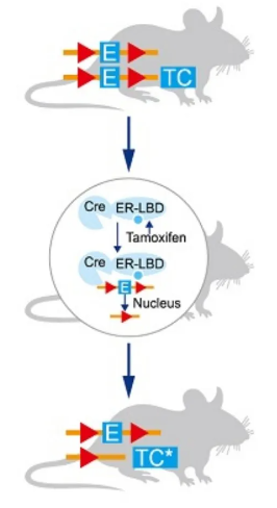
The Split-Cre system divides Cre recombinase into two inactive parts (CreN and CreC), each controlled by different induction conditions. Cre recombinase is only activated and functional when both parts are present and combined. The IBIST system follows a similar principle.
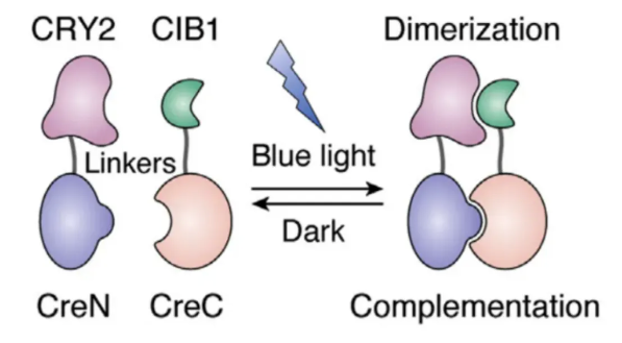
Light-activated Cre (light-inducible Cre) is a type of Cre recombinase that is activated by light. Typically, a light-sensitive protein is fused with Cre, and under specific wavelengths of light, the light-sensitive protein changes its conformation, thereby activating Cre.
In the face of increasingly refined research, Cre recombinase no longer meets all needs. The development of other recombinase systems continues to emerge.
In 1985, Xu-Dong Zhu et al. reported the FLP recombinase derived from Saccharomyces cerevisiae. It is a monomeric protein composed of 423 amino acids that specifically recognizes the FRT (Flp recognition target) site. The FRT site is very similar to the loxP site, consisting of two 13 bp inverted repeat sequences and an 8 bp spacer sequence, with the spacer sequence determining the orientation of the FRT site. The recombination mechanism of the Flp-FRT system is similar to that of the Cre-loxP system.
Many studies have shown the significant recombination efficiency of Cre recombinase, whereas Flp recombinase efficiency is much lower. Buchholz F et al. found that the optimal temperature for Cre recombinase is 37°C, while for Flp recombinase it is 30°C. Although Raymond CS developed a mutant Flp recombinase, Flpo, with thermal stability, narrowing the gap with Cre recombinase, it still falls short of expectations.
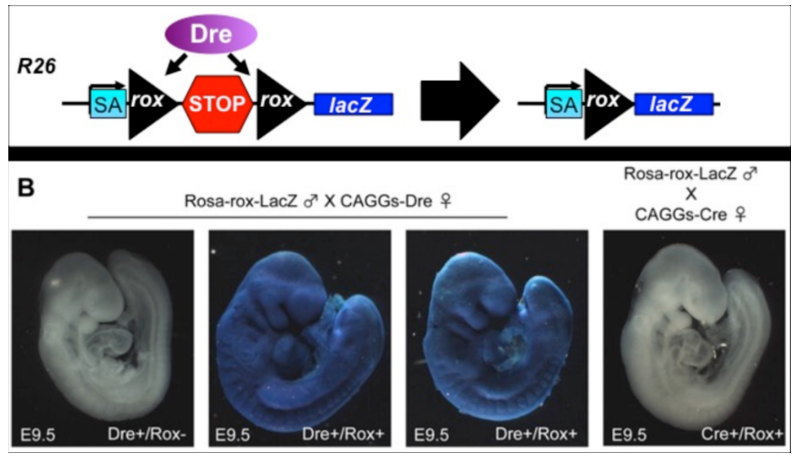
Therefore, in 2009, Brian Sauer et al. cloned the Dre recombinase from the D6 bacteriophage, which can effectively recognize the Rox sequence and mediate DNA recombination. The Rox sequence has a similar function and structure to the loxP sequence but with slight differences; the Rox sequence consists of two 14 bp inverted repeat sequences and a 4 bp spacer sequence. Additionally, the Cre-loxP and Dre-Rox systems are mutually exclusive, allowing them to be used simultaneously.
The development of the Dre system has solved some issues, but the Rox site lacks corresponding mutants, limiting its applications. In 2011, Emiko Suzuki and Manabu Nakayama screened VCre and SCre from Cre recombinase homologs and constructed VloxP/Vlox2272 and SloxP/Slox2272 for them. The VCre/VloxP and SCre/SloxP systems have lower homology with the Cre/loxP system and do not interfere with each other, allowing for combined use.
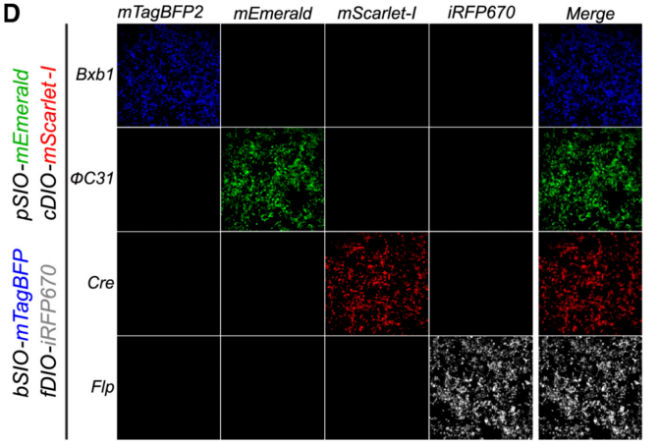
In 2023, Byung Kook Lim's team designed the ΦC31/pSIO system and Bxb1/bSIO system based on the previously reported φC31 and Bxb1, and their specific recognition sites attB, attP, and attL, attR. The ΦC31/pSIO system can also target specific cell types without cross-reacting with Cre and Flp recombinases, further enriching the toolbox of recombinases.
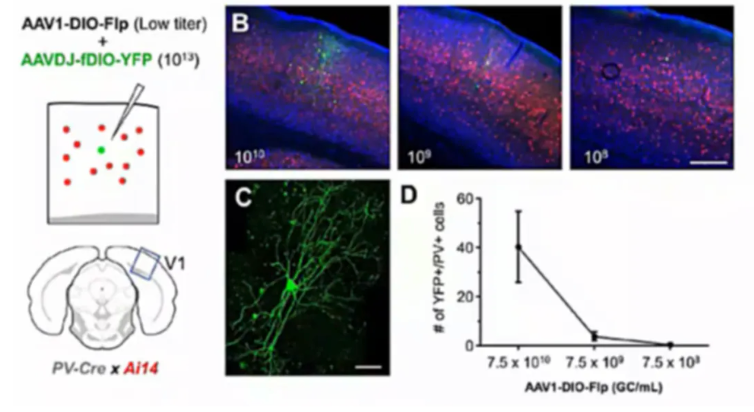
By controlling the mixing ratio of AAV-DIO-Flp and AAV-FDIO-EYFP, the number of neurons can be controlled to achieve sparse and highly bright labeling. The team led by Xu Fuqiang at the Shenzhen Institute of Advanced Technology.
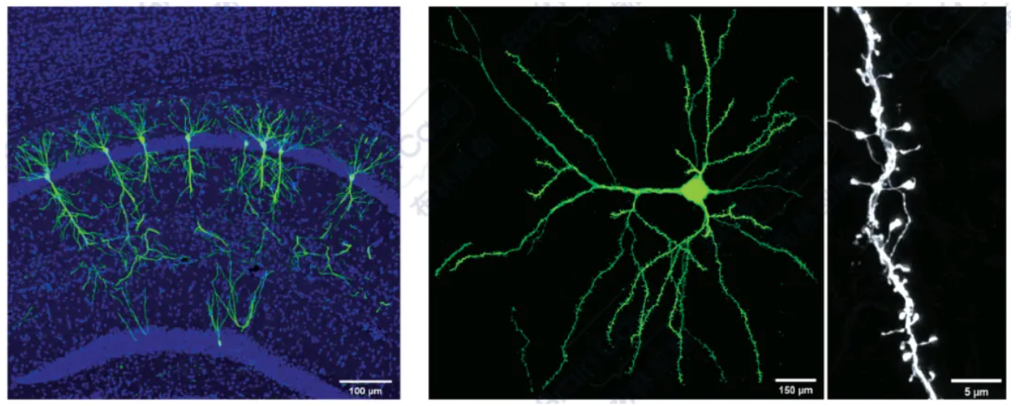
They further optimized this method by adopting a cocktail packaging strategy, co-packaging pAAV-DIO-Flp and pAAV-FDIO-EYFP together, which reduced the issue of mutual exclusion during mixed co-infection of different batches of viruses.
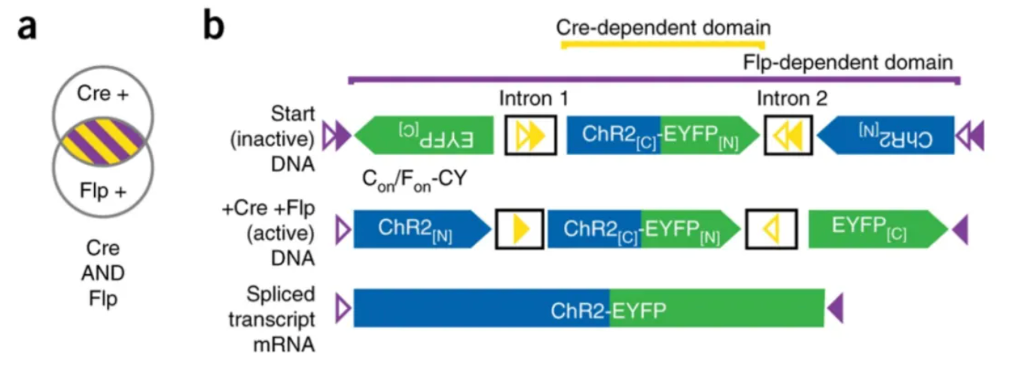
Using Boolean logic principles to modify and synthesize AAV vectors forms a Cre and Flp dual-recombinase cascade control system. This technology overcomes the limitation of single recombinase control systems, which can only study the function and circuit structure of a specific type of neuron. It enables further classification and subclass labeling of specific neurons, offering more selectivity.
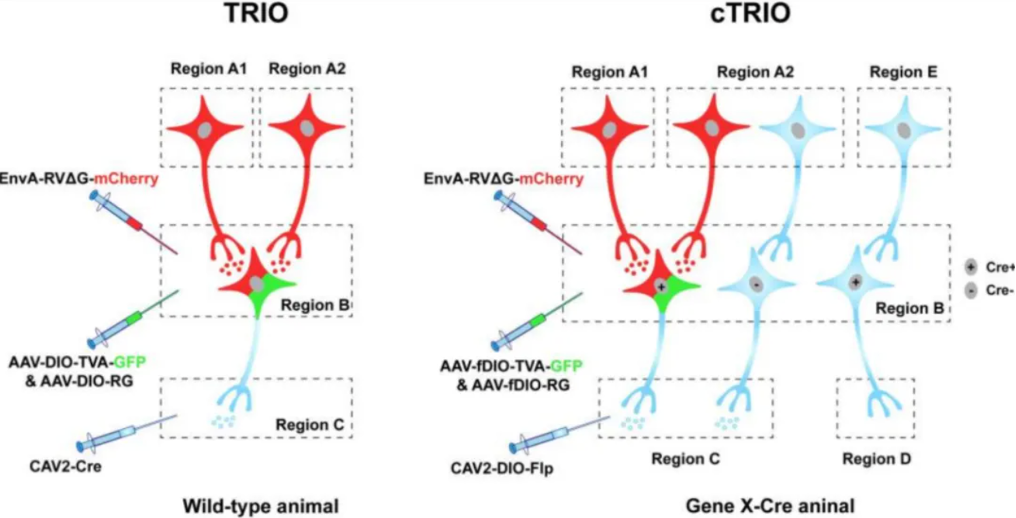
In the cTRIO system, by injecting the retrograde tracer CAV2-DIO-Flp (now more often using the more efficient AAV2-retro-DIO-Flp) into brain area C, neurons in the B→C circuit carry Flp recombinase. This allows for the expression of previously injected AAV-FDIO-TVA-GFP and AAV-FDIO-RVG in brain area B. Finally, by injecting RV-EnvA-ΔG-mCherry, retrograde labeling to the cell bodies in upstream brain areas is achieved, thus marking the A→B→C three-level circuit in Cre mice.